

In the maputils tutorial we show many examples with Python code and figures to illustrate the functionality and flexibility of this module. The documentation below is restricted to the module’s classes and methods.
One of the goals of the Kapteyn Package is to provide a user/programmer basic tools to make plots (with WCS annotation) of image data from FITS files. These tools are based on the functionality of PyFITS and Matplotlib. The methods from these packages are modified in maputils for an optimal support of inspection and presentation of astronomical image data with easy to write and usually very short Python scripts. To illustrate what can be done with this module, we list some steps you need in the process to create a hard copy of an image from a FITS file:
Then for the display:
Of course there are many programs that can do this job some way or the other. But most probably no program does it exactly the way you want or the program does too much. Also many applications cannot be extended, at least not as simple as with the building blocks in maputils.
Module maputils is also very useful as a tool to extract and plot data slices from data sets with more than two axes. For example it can plot so called Position-Velocity maps from a radio interferometer data cube with channel maps. It can annotate these plots with the correct WCS annotation using information about the ‘missing’ spatial axis.
To facilitate the input of the correct data to open a FITS image, to specify the right data slice or to set the pixel limits for the image axes, we implemented also some helper functions. These functions are primitive (terminal based) but effective. You can replace them by enhanced versions, perhaps with a graphical user interface.
Here is an example of what you can expect. We have a three dimensional data set on disk called ngc6946.fits with axes RA, DEC and VELO. The program prompts the user to enter image properties like data limits, axes and axes order. The image below is a data slice in RA, DEC at VELO=50. We changed interactively the color map (keys page-up/page-down) and the color limits (pressing right mouse button while moving the mouse) and saved a hard copy on disk.
In the next code we use keyword parameter promptfie a number of times. Abbreviation ‘fie’ stands for Function Interactive Environment.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 | #!/usr/bin/env python
from kapteyn import wcsgrat, maputils
from matplotlib import pylab as plt
# Create a maputils FITS object from a FITS file on disk
fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
fitsobject.set_imageaxes(promptfie=maputils.prompt_imageaxes)
fitsobject.set_limits(promptfie=maputils.prompt_box)
fitsobject.set_skyout(promptfie=maputils.prompt_skyout)
clipmin, clipmax = maputils.prompt_dataminmax(fitsobject)
# Get connected to Matplotlib
fig = plt.figure()
frame = fig.add_subplot(1,1,1)
# Create an image to be used in Matplotlib
annim = fitsobject.Annotatedimage(frame, clipmin=clipmin, clipmax=clipmax)
annim.Image()
annim.Graticule()
annim.plot()
annim.interact_toolbarinfo()
annim.interact_imagecolors()
annim.interact_writepos()
plt.show()
|
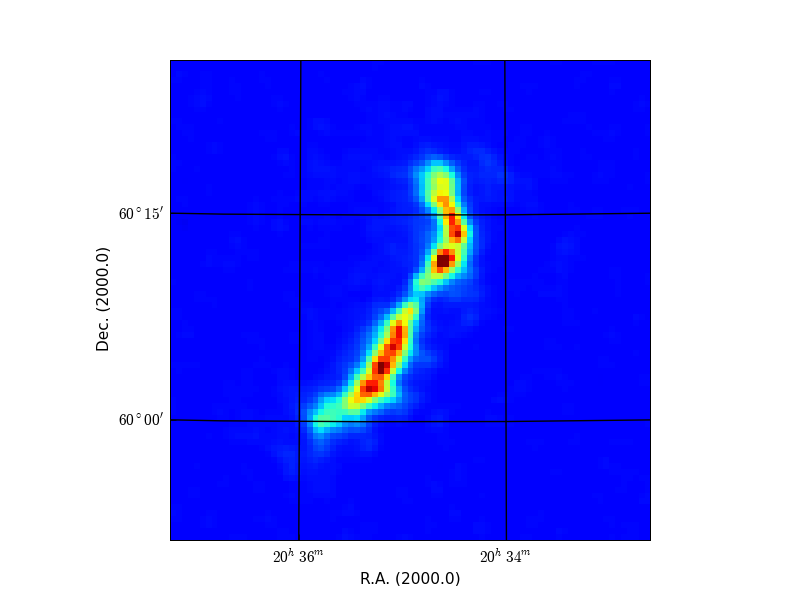
Image from FITS file with graticules and WCS labels
Object from class Colmaplist which has attribute colormaps which is a sorted list with names of colormaps. It has attributes:
- colormaps – List with names of color lookup tables
- cmap_default – Color map set by default to ‘jet’
| Example: |
|
|---|
An external helper function for the FITSimage class to prompt a user to open the right Header and Data Unit (hdu) of a FITS file. A programmer can supply his/her own function of which the return value is a sequence containing the hdu list, the header unit number, the filename and a character for the alternate header.
| Parameters: |
|
|---|---|
| Prompts: |
|
| Returns: |
|
| Notes: | – |
| Examples: | Besides file names of files on disk, PyFITS allows url’s and gzipped files to retrieve FITS files e.g.: http://www.atnf.csiro.au/people/mcalabre/data/WCS/1904-66_ZPN.fits.gz
|
Helper function for FITSimage class. It is a function that requires interaction with a user. Therefore we left it out of any class definition. so that it can be replaced by any other function that returns the position of the data slice in a FITS file.
It prompts the user for the names of the axes of the wanted image. For a 2D FITS data set there is nothing to ask, but for dimensions > 2, we should prompt the user to enter two image axes. Then also a list with pixel positions should be returned. These positions set the position of the data slice on the axes that do not belong to the image. Only with this information the right slice can be extracted.
The user is prompted in a loop until a correct input is given. If a spectral axis is part of the selected image then a second prompt is prepared for the input of the required spectral translation.
| Parameters: |
|
|---|---|
| Prompts: |
|
| Returns: | Tuple with three elements:
|
| Example: | Interactively set the axes of an image using a prompt function: # Create a maputils FITSimage object from a FITS file on disk
fitsobject = maputils.FITSimage('rense.fits')
fitsobject.set_imageaxes(promptfie=maputils.prompt_imageaxes)
|
External helper function which returns the limits in pixels of the x- and y-axis. The input syntax is: xlo,xhi, ylo,yhi. For x and y the names of the image axes are substituted. Numbers can be separated by comma’s and or spaces. A number can also be specified with an expression e.g. 0, 10, 10/3, 100*numpy.pi. All these numbers are converted to integers.
| Parameters: |
|
|---|---|
| Prompts: | Enter pixel limits in Xlo,Xhi, Ylo,Yhi ..... [xlo,xhi, ylo,yhi]: The default should be the axis limits as defined in the FITS header in keywords NAXISn. In a real case this could look like: Enter pixel limits in RAlo,RAhi, DEClo,DEChi ..... [1, 100, 1, 100]: |
| Returns: | Tuple with two elements pxlim, pylim (see parameter description) |
| Notes: | This function does not check if the limits are within the index range of the (FITS)image. This check is done in the FITSimage.set_limits() method of the FITSimage class. |
| Examples: | Use of this function as prompt function in the FITSimage.set_limits() method: fitsobject = maputils.FITSimage('rense.fits')
fitsobject.set_imageaxes(1,2, slicepos=30) # Define image in cube
fitsobject.set_limits(promptfie=maputils.prompt_box)
This ‘box’ prompt needs four numbers. The first is the range in x and the second is the range in y. The input are pixel coordinates, e.g.: >>> 0, 10 10/3, 100*numpy.pi
Note the mixed use of spaces and comma’s to separate the numbers. Note also the use of NumPy for mathematical functions. The numbers are truncated to integers. |
Ask user to enter spectral translation if one of the axes is spectral.
| Parameters: | fitsobj (Instance of class FITSimage) – An object from class FITSimage. From this object we derive the allowed spectral translations. |
|---|---|
| Prompts: | The spectral translation if one of the image axes is a spectral axis.
|
| Returns: |
|
| Example: | >>> fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
>>> print fitsobject.str_spectrans() # Print a list with options first
>>> fitsobject.set_spectrans(promptfie=maputils.prompt_spectrans)
|
Ask user to enter the output sky system if the data is a spatial map.
| Parameters: | fitsobj (Instance of class FITSimage) – An object from class FITSimage. This prompt function uses this object to get information about the axis numbers of the spatial axes in a data structure. |
|---|---|
| Returns: |
|
| Example: | >>> fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
>>> fitsobject.set_skyout(promptfie=maputils.prompt_skyout)
|
Ask user to enter one or two clip values. If one clip level is entered then in display routines the data below this value will be clipped. If a second level is entered, then all data values above this level will also be filtered.
| Parameters: | fitsobj (Instance of class FITSimage) – An object from class FITSimage. |
|---|---|
| Returns: |
|
| Example: | >>> fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
>>> clipmin, clipmax = maputils.prompt_dataminmax(fitsobject)
>>> annim = fitsobject.Annotatedimage(frame, clipmin=clipmin, clipmax=clipmax)
|
Transform a FITS header (read with PyFITS) into a Python dictionary. This is useful if one wants to iterate over all keys in the header. The PyFITS header is not iterable.
Formula for distance on sphere accurate over entire sphere (Vincenty, Thaddeus, 1975). Input and output are in degrees.
| Parameters: |
|
|---|---|
| Examples: | >>> from kapteyn.maputils import dist_on_sphere
>>> print dist_on_sphere(0,0, 20,0)
20.0
>>> print dist_on_sphere(0,30, 20,30)
17.2983302106
|
Usually in a script with only one object of class maputils.Annotatedimage one plots this object, and its derived objects, with method maputils.Annotatedimage.plot(). Matplotlib must be instructed to do the real plotting with pyplot’s function show(). This function does this all.
| Examples: |
|
|---|
This class extracts 2D image data from FITS files. It allows for external functions to prompt users for relevant input like the name of the FITS file, which header in that file should be used, the axis numbers of the image axes, the pixel limits and a spectral translation if one of the selected axes is a spectral axis. All the methods in this class that allow these external functions for prompting can also be used without these functions. Then one needs to know the properties of the FITS data beforehand.
| Parameters: |
|
||
|---|---|---|---|
| Attributes: |
|
||
| Notes: | The object is initialized with a default position for a data slice if the dimension of the FITS data is > 2. This position is either the value of CRPIX from the header or 1 if CRPIX is outside the range [1, NAXIS]. Values -inf and +inf in a dataset are replaced by NaN’s (not a number number). We know that Matplotlib’s methods have problems with these values, but these methods can deal with NaN’s. |
||
| Examples: | PyFITS allows URL’s to retrieve FITS files. It can also read gzipped files e.g.:
Use Maputil’s prompt function prompt_fitsfile() to get user interaction for the FITS file specification. >>> fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
|
||
| Methods: |
A FITS file can contain a data set of dimension n. If n < 2 we cannot display the data without more information. If n == 2 the data axes are those in the FITS file, Their numbers are 1 and 2. If n > 2 then we have to know the numbers of those axes that are part of the image. For the other axes we need to know a pixel position so that we are able to extract a data slice.
Attribute dat is then always a 2D array.
| Parameters: |
|
|---|---|
| Raises: |
|
A dictionary with objects from class FITSaxis. One object for each axis. The dictionary keys are the axis numbers. See also second example at method FITSaxis.printattr().
A list with strings representing the spectral translations that are possible for the current image axis selection.
The selected spectral translation
One or a list with integers that represent pixel positions on axes in the data set that do not belong to the image. At these position, a slice with image data is extracted.
Image data from the selected FITS file. It is always a 2D data slice and its size can be found in attribute imshape.
The shape of the array map.
Images with only one spatial axis, need another spatial axis to produces useful world coordinates. This attribute is extracted from the relevant axis in attribute slicepos.
An object from class Projection as defined in wcs.
The axis numbers corresponding with the X-axis and Y-axis in the image.
| Note: | The aspect ratio is reset (to None) after each call to this method. |
|---|---|
| Examples: | Set the image axes explicitly: >>> fitsobject = maputils.FITSimage('rense.fits')
>>> fitsobject.set_imageaxes(1,2, slicepos=30)
Set the images axes in interaction with the user using a prompt function: >>> fitsobject = maputils.FITSimage('rense.fits')
>>> fitsobject.set_imageaxes(promptfie=maputils.prompt_imageaxes)
Enter (part of) the axis names. Note the minimal matching and case insensitivity. >>> fitsobject = maputils.FITSimage('rense.fits')
>>> fitsobject.set_imageaxes('ra','d', slicepos=30)
|
This method sets the image box. That is, it sets the limits of the image axes in pixels. This can be a useful feature if one knows which part of an image contains the interesting data.
| Parameters: |
|
|---|---|
| Examples: | Ask user to enter limits with prompt function prompt_box() >>> fitsobject = maputils.FITSimage('rense.fits')
>>> fitsobject.set_imageaxes(1,2, slicepos=30) # Define image in cube
>>> fitsobject.set_limits(promptfie=maputils.prompt_box)
|
Set spectral translation or ask user to enter a spectral translation if one of the axes in the current FITSimage is spectral.
| Parameters: |
|
|---|---|
| Examples: | Set a spectral translation using 1) a prompt function, 2) a spectral translation for which we don’t know the code for the conversion algorithm and 3) set the translation explicitly: >>> fitsobject.set_spectrans(promptfie=maputils.prompt_spectrans)
>>> fitsobject.set_spectrans(spectrans="VOPT-???")
>>> fitsobject.set_spectrans(spectrans="VOPT-V2W")
|
Set the output sky definition. Mouse positions and coordinate labels will correspond to the selected definition. The method will only work if both axes are spatial axes.
| Parameters: |
|
|---|---|
| Notes: | The method sets an output system only for data with two spatial axes. For XV maps the output sky system is always the same as the native system. |
This method couples the data slice that represents an image to a Matplotlib Axes object (parameter frame). It returns an object from class Annotatedimage which has only attributes relevant for Matplotlib.
| Parameters: |
|
||
|---|---|---|---|
| Attributes: | See documentation at Annotatedimage |
||
| Returns: | An object from class Annotatedimage |
||
| Examples: | >>> f = maputils.FITSimage("ngc6946.fits")
>>> f.set_imageaxes(1, 3, slicepos=51)
>>> annim = f.Annotatedimage()
or:
|
Return the aspect ratio of the pixels in the current data structure defined by the two selected axes. The aspect ratio is defined as pixel height / pixel width.
| Example: | >>> fitsobject = maputils.FITSimage('m101.fits')
>>> print fitsobject.get_pixelaspectratio()
1.0002571958
|
|---|---|
| Note: | If a header has only a cd matrix and no values for CDELT, then these values are set to 1. This gives an aspect ratio of 1. |
Usually a user will set the figure size manually with Matplotlib’s figure(figsize=...) construction. For many plots this is a waste of white space around the plot. This can be improved by taking the aspect ratio into account and adding some extra space for labels and titles. For aspect ratios far from 1.0 the number of pixels in x and y are taken into account.
A handy feature is that you can enter the two values in centimeters if you set the flag cm to True.
If you have a plot which is higher than its width and you want to fit in on a A4 page then use:
>>> f = maputils.FITSimage(externalheader=header)
>>> figsize = f.get_figsize(ysize=21, cm=True)
>>> fig = plt.figure(figsize=figsize)
>>> frame = fig.add_subplot(1,1,1)
Print the meta information from the selected header. Omit items of type HISTORY. It prints both real FITS headers and headers given by a dictionary.
| Returns: | A string with the header keywords |
||
|---|---|---|---|
| Examples: | If you think a user needs more information from the header than can be provided with method str_axisinfo() it can be useful to display the contents of the selected FITS header. This is the entire header and not a selected alternate header.
|
For each axis in the FITS header, return a string with the data related to the World Coordinate System (WCS).
| Parameters: |
|
||
|---|---|---|---|
| Returns: | A string with WCS information for each axis in axnum. |
||
| Examples: | Print useful header information after the input of the FITS file and just before the specification of the image axes:
Print extended information for the two image axes only: >>> print str_axisinfo(axnum=fitsobject.axperm, long=True)
|
||
| Notes: | For axis numbers outside the range of existing axes in the FITS file, nothing will be printed. No exception will be raised. |
Compose a string with information about the data related to the current World Coordinate System (WCS) (e.g. which axes are longitude, latitude or spectral axes)
| Returns: | String with WCS information for the current Projection object. |
||
|---|---|---|---|
| Examples: | Print information related to the world coordinate system:
|
Compose a string with the possible spectral translations for this data.
| Returns: | String with information about the allowed spectral translations for the current Projection object. |
|---|---|
| Examples: | Print allowed spectral translations: >>> print fitsobject.str_spectrans()
|
Get minimum and maximum value of data in entire data structure defined by the current FITS header or in a slice. These values can be important if you want to compare different images from the same source (e.g. channel maps in a radio data cube).
| Parameters: | box (Boolean) – Find min, max in data or if set to True in data slice (with limits). |
||
|---|---|---|---|
| Returns: | min, max, two floating point numbers representing the minimum and maximum data value in data units of the header (BUNIT). |
||
| Note: | We assume here that the data is read when a FITSobject was created. Then the data is filtered and the -inf, inf values are replaced by NaN’s. |
||
| Example: | Note the difference between the min, max of the entire data or the min, max of the slice (limited by a box):
|
Given the pixel coordinates of a slice, return the world coordinates of these pixel positions and their units. For example in a 3-D radio cube with axes RA-DEC-FREQ one can have several RA-DEC images as function of FREQ. This FREQ is given in pixels coordinates in attribute slicepos. The world coordinates are calculated using the Projection object which is also an attribute.
| Parameters: |
|
|---|---|
| Returns: | A tuple with two elements: world and units. Element world is either an empty list or a list with one or more world coordinates. The number of coordinates is equal to the number of axes in a data set that do not belong to the extracted data which can be a slice. For each world coordinate there is a unit in element units. |
| Note: | This method first calculates a complete set of world coordinates. Where it did not define a slice position, it takes the header value CRPIXn. So if a map is defined with only one spatial axes and the missing spatial axis is found in slicepos than we have two matching pixel coordinates for which we can calculate world coordinates. So by definition, if a slice is a function of a spatial coordinate, then its world coordinate is found by using the matching pixel coordinate which, in case of a spatial map, corresponds to the projection center. |
| Example: | >>> vel, uni = fitsobj.slice2world(spectra="VOPT-???")
>>> velinfo = "ch%d = %.1f km/s" % (ch, vel[0]/1000.0)
or: >>> vel, uni = fitsobj.slice2world(spectra=”VOPT-???”, userunits=”km/s”) |
If a header contains PC or CD elements, and not all the ‘classic’ elements for a WCS then a number of FITS readers could have a problem if they don’t recognize a PC and CD matrix. What can be done is to derive the missing header items, CDELTn and CROTA from these headers and add them to the header.
What is a ‘classic’ FITS header?
(See also http://fits.gsfc.nasa.gov/fits_standard.html) For the transformation between pixel coordinates and world coordinates, FITS supports three conventions. First some definitions:
An intermediate pixel coordinate \(q_i\) is calculated from a pixel coordinates \(p\) with:
Rj are the pixel coordinate elements of a reference point (FITS header item CRPIXj), j is an index for the pixel axis and i for the world axis The matrix \(m_{ij}\) must be non-singular and its dimension is NxN where N is the number of world coordinate axes (given by FITS header item NAXIS).
The conversion of \(q_i\) to intermediate world coordinate \(x_i\) is a scale \(s_i\):
Formalism 1 (PC keywords)
Formalism 1 encodes \(m_{ij}\) in so called PCi_j keywords and scale factor \(s_i\) are the values of the CDELTi keywords from the FITS header.
It is obvious that the value of CDELT should not be 0.0.
Formalism 2 (CD keywords)
If the matrix and scaling are combined we get for the intermediate WORLD COORDINATE \(x_i\):
FITS keywords CDi_j encodes the product \(s_i m_{ij}\). The units of \(x_i\) are given by FITS keyword CTYPEi.
Formalism 3 (Classic)
This is the oldest but now deprecated formalism. It uses CDELTi for the scaling and CROTAn for a rotation of the image plane. n is associated with the latitude axis so often one sees CROTA2 in the header if the latitude axis is the second axis in the dataset
Following the FITS standard, a number of rules is set:
- CDELT and CROTA may co-exist with the CDi_j keywords but must be ignored if an application supports the CD formalism.
- CROTAn must not occur with PCi_j keywords
- CRPIXj defaults to 0.0
- CDELT defaults to 1.0
- CROTA defaults to 0.0
- PCi_j defaults to 1 if i==j and to 0 otherwise. The matrix must not be singular
- CDi_j defaults to 0.0. The matrix must not be singular.
- CDi_j and PCi_j must not appear together in a header.
Alternate WCS axis descriptions
A World Coordinate System (WCS) can be described by an alternative set of keywords. For this keywords a character in the range [A..Z] is appended. In our software we made the assumption that the primary description contains all the necessary information to derive a ‘classic’ header and therefore we will ignore alternate header descriptions.
Conversion to a formalism 3 (‘classic’) header
Many FITS readers from the past are not upgraded to process FITS files with headers written using formalism 1 or 2. The purpose of this application is to convert a FITS file to a file that can be read and interpreted by old FITS readers. For GIPSY we require FITS headers to be written using formalism 3. If keywords are missing, they will be derived and a comment will be inserted about the keyword not being an original keyword.
The method that converts the header, tries to process it first with WCSLIB (tools to interpret the world coordinate system as described in a FITS header). If this fails, then we are sure that the header is incorrect and we cannot proceed. One should use a FITS tool like ‘fv’ (the Interactive FITS File Editor from Nasa) to repair the header.
The conversion process starts with exploring the spatial part of the header. It knows which axes are spatial and it reads the corresponding keywords (CDELTi, CROTAn, CDi_j, PCi_j and PC00i_00j (old format for PC elements). If there is no CD or PC matrix, then the conversion routine returns the unaltered original header. If it finds a PC matrix and no CD matrix then the header should contain CDELT keywords. With the values of these keywords we create a CD matrix:
Notes:
- We replaced notation i_j by ij so cd11 == CD1_1
- For the moment we restricted the problem to the 2 dim. spatial case because that is what we need to retrieve a value for CROTA, the rotation of the image.)
- We assumed that the PC matrix did not represent transposed axes as in:
If cd12 == 0.0 and cd12 == 0.0 then CROTA is obviously 0. There is no rotation and CDELT1 = cd11, CDELT2 = cd22
If one or both values of cd12, cd21 is not zero then we expect a value for CROTA unequal to zero and/or skew.
We calculate the scaling parameters CDELT with:
CDELT1 = sqrt(cd11*cd11+cd21*cd21)
CDELT2 = sqrt(cd12*cd12+cd22*cd22)
The determinant of the matrix is:
det = cd11*cd22 - cd12*cd21
This value cannot be 0 because we required that the matrix is non-singular. Further we distinguish two situations: a determinant < 0 and a determinant > 0 (zero was already excluded). Then we derive two rotations. If these are equal, the image is not skewed. If they are not equal, we derive the rotation from the average of the two calculated angles. As a measure of skew, we calculated the difference between the two rotation angles. Here is a piece of the actual code:
1 2 3 4 5 6 7 8 9 | sign = 1.0
if det < 0.0:
cdeltlon_cd = -cdeltlon_cd
sign = -1.0
rot1_cd = atan2(-cd21, sign*cd11)
rot2_cd = atan2(sign*cd12, cd22)
rot_av = (rot1_cd+rot2_cd)/2.0
crota_cd = degrees(rot_av)
skew = degrees(abs(rot1_cd-rot2_cd))
|
New values of CDELT and CROTA will be inserted in the new ‘classic’ header only if they were not part of the original header.
The process continues with non spatial axes. For these axes we cannot derive a rotation. We only need to find a CDELTi if for axis i no value could be found in the header. If this value cannot be derived from the a CD matrix (usually with diagonal element CDi_i) then the default 1.0 is assumed. Note that there will be a warning about this in the comment string for the corresponding keyword in the new ‘classic’ FITS header.
Finally there is some cleaning up. First all CD/PC elements are removed for all the axes in the data set. Second, some unwanted keywords are removed. The current list is: [“XTENSION”, “EXTNAME”, “EXTEND”]
See also: Calabretta & Greisen: ‘Representations of celestial coordinates in FITS’, section 6
| Parameters: | - – No parameters |
||
|---|---|---|---|
| Returns: | A tuple with three elements:
|
||
| Notes: | This method is tested with FITS files:
|
||
| Example: |
|
The current FITSimage object must contain a number of spatial maps. This method then reprojects these maps so that they conform to the input header.
Imagine an image and a second image of which you want to overlay contours on the first one. Then this method uses the current data to reproject to the input header and you will end up with a new FITSimage object which has the spatial properties of the input header and the reprojected data of the current FITSimage object.
Also more complicated data structures can be used. Assume you have a data cube with axes RA, Dec and Freq. Then this method will reproject all its spatial subsets to the spatial properties of the input header.
The current FITSimage object tries to keep as much of its original FITS keywords. Only those related to spatial data are copied from the input header. The size of the spatial map can be limited or extended. The axes that are not spatial are unaltered.
The spatial information for both data structures are extracted from the headers so there is no need to specify the spatial parts of the data structures.
The image that is reprojected can be limited in size with parameters pxlim_dst and pylim_dst. If the input is a FITSimage object, then these parameters (pxlim_dst and pylim_dst) are copied from the axes lengths set with method set_limits() for that FITSimage object.
| Parameters: |
|
||||||
|---|---|---|---|---|---|---|---|
| Examples: | -Set limits for axes outside the spatial map. Assume a data structure with axes RA-DEC-FREQ-STOKES for which the RA-DEC part is reprojected to a set RA’-DEC’-FREQ-STOKES. The ranges for FREQ and STOKES set the number of spatial maps in this data structure. One can limit these ranges with plimlo and plimhi.
Note that if the data structure was represented by axes FREQ-RA-STOKES-DEC then the examples above are still valid because these set the limits on the repeat axes FREQ and POL whatever the position of these axes in the data structure. -Use and modify the current header to change the data. The example shows how to rotate an image and display the result.
-Use an external header and change keywords in that header before the re-projection: >>> Rotfits = Basefits.reproject_to(externalheader,
naxis1=800, naxis2=800,
crpix1=400, crpix2=400)
-Use the FITS file’s own header. Change it and use it as an external header
|
||||||
| Tests: |
|
Note
If you want to align an image with the direction of the north, then the value of CROTAn (e.g. CROTA2) should be set to zero. To ensure that the data will be rotated, use parameter rotation with a dummy value so that the header used for the re-projection is a ‘classic’ header:
e.g.:
>>> Rotfits = Basefits.reproject_to(rotation=0.0, crota2=0.0)
Warning
Values for CROTAn in parameter fitskeys overwrite values previously set with keyword rotation.
Warning
Changing values of CROTAn will not always result in a rotated image. If the world coordinate system was defined using CD or PC elements, then changing CROTAn will only add the keyword but it is never read because CD & PC transformations have precedence.
This method copies current data and current header to a FITS file on disk. This is useful if either header or data comes from an external source. If no file name is entered then a file name will be composed using current date and time of writing. The name then start with ‘FITS’.
| Parameters: |
|
||
|---|---|---|---|
| Raises: |
|
||
| Examples: | Artificial header and data:
|
This is one of the core classes of this module. It sets the connection between the FITS data (created or read from file) and the routines that do the actual plotting with Matplotlib. The class is usually used in the context of class FITSimage which has a method that prepares the parameters for the constructor of this class.
| Parameters: |
|
|---|---|
| Attributes: |
|
| Methods: |
Matplotlib scales image values between 0 and 1 for its distribution of colors from the color map. With this method we set the image values which we want to scale between 0 and 1. The default image values are the minimum and maximum of the data in Annotatedimage.data. If you want to inspect a certain range of data values you need more colors in a smaller intensity range, then use different clipmin and clipmax in the constructor of Annotatedimage or in this method.
| Parameters: |
|
|---|---|
| Examples: | >>> fitsobj = maputils.FITSimage("m101.fits")
>>> annim = fitsobj.Annotatedimage(frame, cmap="spectral")
>>> annim.Image(interpolation='spline36')
>>> annim.set_norm(10000, 15500)
or: >>> fitsobj = maputils.FITSimage("m101.fits")
>>> annim = fitsobj.Annotatedimage(frame, cmap="spectral", clipmin=10000, clipmax=15500)
|
| Notes: | It is also possible to change the norm after an image has been displayed. This enables a programmer to setup key interaction for changing the clip levels in an image for example when the default clip levels are not suitable to inspect a certain data range. Usually the color editing (with Annotatedimage.interact_imagecolors()) can do this job very well so we think there is not much demand in a scripting environment. With GUI’s it will be different. |
Method to set the initial color map for images, contours and colorbars. These color maps are either strings (e.g. ‘jet’ or ‘spectral’) from a list with Matplotlib color maps or it is a path to a color map on disk (e.g. cmap=”/home/user/luts/mousse.lut”). If the color map is not found in the list with known color maps then it is added to the list, which is a global variable called cmlist. (see: maputils.cmlist)
The Kapteyn Package has also a list with useful color maps. See example below or example ‘mu_luttest.py’ in the Tutorial maputils module.
If you add the event handler interact_imagecolors() it is possible to change colormaps with keyboard keys and mouse.
| Parameters: |
|
|---|---|
| Examples: | >>> fitsobj = maputils.FITSimage("m101.fits")
>>> annim = fitsobj.Annotatedimage(frame, clipmin=10000, clipmax=15500)
>>> annim.set_colormap("spectral")
or use the constructor as in: >>> annim = fitsobj.Annotatedimage(frame, cmap="spectral", clipmin=10000, clipmax=15500)
Get extra lookup tables from Kapteyn Package (by default, these luts are appended at creation time of cmlist, see maputils.cmlist) >>> extralist = mplutil.VariableColormap.luts()
>>> maputils.cmlist.add(extralist)
|
Method to write current colormap rgb values to file on disk. If you add the event handler interact_imagecolors(), this method is automatically invoked if you press key ‘m’.
This method is only useful if the colormap changes i.e. in an interactive environment.
| Parameters: | filename (Python stringz) – Name of file to which the current (modified) color look up table (lut) is written. This lut can be imported as a new lut in another session. |
|---|
Set the color of bad pixels. If you add the event handler interact_imagecolors(), this method steps through a list of colors for the bad pixels in an image.
| Parameters: |
|
|---|---|
| Example: | >>> annim.set_blankcolor('c')
|
Set the aspect ratio. Overrule the default aspect ratio which corrects pixels that are not square in world coordinates. Can be useful if you want to stretch images for which the aspect ratio doesn’t matter (e.g. XV maps).
| Parameters: | aspect (Float) – The aspect ratio is defined as pixel height / pixel width. With this value one can stretch an image in x- or y direction. The default is such that 1 arcmin in x has the same length in cm as 1 arcmin in y. |
|---|---|
| Example: | >>> annim = fitsobj.Annotatedimage(frame)
>>> annim.set_aspectratio(1.2)
|
This method compiles and returns a help text for color map interaction.
Setup of an image. This method is a call to the constructor of class Image with a default value for most of the keyword parameters. This created object has an attribute im which is an instance of Matplotlib’s imshow() method. This object has a plot method. This method is used by the more general Annotatedimage.plot() method.
| Parameters: | kwargs (Python keyword parameters) – From the documentation of Matplotlib we learn that for method imshow() (used in the plot method if an Image) a few interesting keyword arguments remain:
|
|---|---|
| Attributes: |
The object generated after a call to Matplotlib’s imshow(). |
| Example: | >>> fitsobject = maputils.FITSimage(promptfie=maputils.prompt_fitsfile)
>>> annim = fitsobject.Annotatedimage()
>>> annim.Image(interpolation="spline36")
or create an image but make it invisible: >>> annim.Image(visible=False)
|
Matplotlib’s method imshow() is able to produce RGB images. To create a real RGB image, we need three arrays with identical shape representing the red, green and blue components. Method imshow() requires data scaled between 0 and 1.
This utility method prepares a composed and scaled data array derived from three FITSimage objects. It scales the composed array and not the individual image arrays. The method allows for a function or lambda expression to be entered to process the scaled data. The world coordinate system (e.g. to plot graticules) is copied from the current Annotatedimage object. Note that for the three images only the shape of the array must be equal to the shape of the data of the current Annotatedimage object.
| Parameters: |
|
||
|---|---|---|---|
| Note: | A RGB image does not interact with a colormap. Interacting with a colormap (e.g. after adding annim.interact_imagecolors() in the example below) is not forbidden but it gives weird results. To rescale the data, for instance for a better view of interesting data, you need to enter a function or Lambda expression with parameter fun. |
||
| Example: |
|
Setup of contour lines. This method is a call to the constructor of class Contours with a number of default parameters. Either it plots single contour lines or a combination of contour lines and filled regions between the contours. The colors are taken from the current colormap.
| Parameters: |
|
||
|---|---|---|---|
| Methods: |
|
||
| Examples: |
>>> annim.Contours(filled=True)
In the next example note the plural form of the standard Matplotlib keywords. They apply to all contours: >>> annim.Contours(colors='w', linewidths=2)
Set levels and the line style for negative contours: >>> annim.Contours(levels=[-500,-300, 0, 300, 500], negative="dotted")
A combination of keyword parameters with less elements than the number of contour levels: >>> cont = annim.Contours(linestyles=('solid', 'dashed', 'dashdot', 'dotted'),
linewidths=(2,3,4), colors=('r','g','b','m'))
Example of setting of properties for all and 1 contour with setp_contour(): >>> cont = annim.Contours(levels=range(10000,16000,1000))
>>> cont.setp_contour(linewidth=1)
>>> cont.setp_contour(levels=11000, color='g', linewidth=3)
Plot a (formatted) label near a contour with setp_label(): >>> cont2 = annim.Contours(levels=(8000,9000,10000,11000))
>>> cont2.setp_label(11000, colors='b', fontsize=14, fmt="%.3f")
>>> cont2.setp_label(fontsize=10, fmt="$%g \lambda$")
|
This method is a call to the constructor of class Colorbar with a number of default parameters. A color bar is an image which represents the current color scheme. It annotates the colors with image values so that it is possible to get an idea of the distribution of the values in your image.
| Parameters: |
|
||
|---|---|---|---|
| Methods: | Colorbar.set_label() - Plot a title along the long side of the colorbar. |
||
| Examples: | A basic example were the font size for the ticks are set:
Set frames for Image and Colorbar: >>> frame = fig.add_axes((0.1, 0.2, 0.8, 0.8))
>>> cbframe = fig.add_axes((0.1, 0.1, 0.8, 0.1))
>>> annim = fitsobj.Annotatedimage(cmap="Accent", clipmin=8000, frame=frame)
>>> colbar = annim.Colorbar(fontsize=8, orientation='horizontal', frame=cbframe)
Create a title for the colorbar and change its font size: >>> units = r'$ergs/(sec.cm^2)$'
>>> colbar.set_label(label=units, fontsize=24)
|
This method is a call to the constructor of class wcsgrat.Graticule with a number of default parameters.
It calculates and plots graticule lines of constant longitude or constant latitude. The description of the parameters is found in wcsgrat.Graticule. An extra parameter is visible. If visible is set to False than we can plot objects derived from this class such as ‘Rulers’ and ‘Insidelabels’ without plotting unwanted graticule lines and labels.
| Methods: |
|
|---|
Other parameters such as hdr, axperm, pxlim, pylim, mixpix, skyout and spectrans are set to defaults in the context of this method and should not be overwritten.
| Examples: |
Set the range in world coordinates and set the positions for the labels with (X, Y): >>> X = arange(0,360.0,15.0)
>>> Y = [20, 30,45, 60, 75, 90]
>>> grat = annim.Graticule(wylim=(20.0,90.0), wxlim=(0,360),
startx=X, starty=Y)
Add a ruler, based on the current Annotatedimage object: >>> ruler3 = annim.Ruler(23*15,30,22*15,15, 0.5, 1, world=True,
fmt=r"$%4.0f^\prime$",
fun=lambda x: x*60.0, addangle=0)
>>> ruler3.setp_labels(color='r')
Add world coordinate labels inside the plot. Note that these are derived from the current Graticule object. >>> grat.Insidelabels(wcsaxis=0, constval=-51, rotation=90, fontsize=10,
color='r', ha='right')
|
|---|
This method is a call to the constructor of class Pixellabels with a number of default parameters. It sets the annotation along a plot axis to pixel coordinates.
| Parameters: |
|
|---|---|
| Examples: | >>> fitsobject = maputils.FITSimage("m101.fits")
>>> annim = fitsobject.Annotatedimage()
>>> annim.Pixellabels(plotaxis=("top","right"), color="b", markersize=10)
or separate the labeling so that you can give different properties for different axes. In this case we shift the labels along the top axis towards the axis line with va=’top’: >>> annim.Pixellabels(plotaxis='top', va='top')
>>> annim.Pixellabels(plotaxis='right')
|
| Notes: | If a pixel offset is given for this Annimated object, then plot the pixel labels with this offset. |
Drawing minor tick marks is as easy or as difficult as finding the positions of major tick marks. Therefore we decided that the best way to draw minor tick marks is to calculate a new (but invisible) graticule object. Only the tick marks are visible.
| Parameters: |
|
||
|---|---|---|---|
| Notes: | Minor tick marks are also plotted at the positions of major tick marks. By default this will not be visible. It is visible if you user a longer marker size, a different color or a marker with increased width. The default marker size is set to 2. |
||
| Returns: | This method returns a graticule object. Its properties can be changed in the calling environment with the appropriate methods (e.g. wcsgrat.Graticule.setp_tickmark()). |
||
| Examples: |
Adding parameters to change attributes: >>> grat2 = grat.minortickmarks(3, 5, color="#aa44dd", markersize=3, markeredgewidth=2)
Minor tick marks only along x axis: >>> grat2 = minortickmarks(grat, 3, None)
|
Objects from class Beam are graphical representations of the resolution of an instrument. The beam is centered at a position xc, yc. The major axis of the beam is the FWHM of the longest distance between two opposite points on the ellipse. The angle between the major axis and the North is the position angle.
A beam is an ellipse in world coordinates. To draw a beam given the input parameters, points are calculated in world coordinates so that angle and required distance of sample points on the ellipse are correct on a sphere.
| Parameters: |
|
||
|---|---|---|---|
| Examples: |
|
||
| Properties: | A selection of keyword arguments for the beam (which is a Matplotlib Polygon object) are:
|
Plot marker symbols at given positions. This method creates objects from class Marker. The constructor of that class needs positions in pixel coordinates. Here we allow positions to be defined in a string which can contain either world- or pixel coordinates (or a mix of both). If x and y coordinates are known, or read from file, one can also enter this data without parsing. The keyword arguments x and y can be used to enter pixel coordinates or world coordinates.
| Parameters: |
|
||||
|---|---|---|---|---|---|
| Returns: | Object from class Marker |
||||
| Examples: | In the first example we show 4 markers plotted in the projection center (given by header values CRPIX):
In the second example we show how to plot a sequence of markers. Note the use of the different keyword arguments and the role of flag world to force the given values to be processed in pixel coordinates:
In the next example we show how to use method positionsfromfile() in combination with this Marker method to read positions from a file and to plot them. The positions in the file are world coordinates. Method positionsfromfile() returns pixel coordinates: fn = 'smallworld.txt'
xp, yp = annim.positionsfromfile(fn, 's', cols=[0,1])
annim.Marker(x=xp, y=yp, mode='pixels', marker=',', color='b')
|
This method prepares arguments for a call to function rulers.Ruler() in module rulers
Note that this method sets a number of parameters which cannot be changed like projection, mixpix, pxlim, pylim and aspectratio, which are all derived from the properties of the current maputils.Annotatedimage object.
Construct an object that represents an area in the sky. Usually this is an ellipse, rectangle or regular polygon with given center and other parameters to define its size or number of angles and the position angle. The object is plotted in a way that the sizes and angles, as defined on a sphere, are preserved. The objects need a ‘prescription’. This is a recipe to calculate a distance to a center point (0,0) as function of an angle in a linear and flat system. Then the object perimeter is re-calculated for a given center (xc,yc) and for corresponding angles and distances on a sphere.
If prescription=None, then this method expects two arrays lons and lats. These are copied unaltered as vertices for an irregular polygon.
For cylindrical projections it is possible that a polygon in a all sky plot crosses a boundary (e.g. 180 degrees longitude if the projection center is at 0 degrees). Then the object is splitted into two parts one for the region 180-phi and one for the region 180+phi where phi is an arbitrary positive angle. This splitting is done for objects with and without a prescription.
| Parameters: |
|
|---|
Plot all objects stored in the objects list for this Annotated image.
| Example: |
|
|---|
This is a helper method for method wcs.Projection.toworld(). It converts pixel positions from a map to world coordinates. The difference with that method is that this method has its focus on maps, i.e. two dimensional structures. It knows about the missing spatial axis if your data slice has only one spatial axis. Note that pixels in FITS run from 1 to NAXISn and that the pixel coordinate equal to CRPIXn corresponds to the world coordinate in CRVALn.
| Parameters: |
|
||||
|---|---|---|---|---|---|
| Note: | If somewhere in the process an error occurs, then the return values of the world coordinates are all None. |
||||
| Returns: | Three world coordinates: xw which is the world coordinate for the x-axis, yw which is the world coordinate for the y-axis and (if matchspatial=True) missingspatial which is the world coordinate that belongs to the missing spatial axis. If there is not a missing spatial axis, then the value of this output parameter is None. So you don’t need to know the structure of the map beforehand. You can test whether the last value is None or not None in the calling environment. |
||||
| Examples: | We have a test set with:
Now let us try to find the world coordinates of a RA-VELO map at (crpix1, crpix3) at slice position DEC=51. We should get three numbers which are all equal to the value of CRVALn
Or work with a sequence of numbers (list, tuple of NumPy ndarray object) as in this example:
|
This is a helper method for method wcs.Projection.topixel(). It knows about the missing spatial axis if a data slice has only one spatial axis. It converts world coordinates in units (given by the FITS header, or the spectral translation) from a map to pixel coordinates. Note that pixels in FITS run from 1 to NAXISn.
| Parameters: |
|
|---|---|
| Returns: | Two pixel coordinates: x which is the world coordinate for the x-axis and y which is the world coordinate for the y-axis. If somewhere in the proces an error occurs, then the return values of the pixel coordinates are all None. |
| Notes: | This method knows about the pixel on the missing spatial axis (if there is one). This pixel is usually the pixel coordinate of the slice if the dimension of the data is > 2. |
| Examples: | See example at toworld() |
This convenience method belongs to class Annotatedimage which represents a two dimensional map which could be a slice (slicepos) from a bigger data structure and/or could be limited by limits on the pixel ranges of the image axes (pxlim, pylim). Then, for a sequence of coordinates in x and y, return a sequence with Booleans with True for a coordinate within the boundaries of this map and False when it is outside the boundaries of this map. This method can work with either sequences of coordinates (parameters x and y) or a string with a position (parameter pos). If parameters x and y are used then parameter world sets these coordinates to world- or pixel coordinates.
| Parameters: |
|
|---|---|
| Raises: |
|
| Returns: |
|
| Note: | For programmers: note the similarity to method Marker() with respect to the use of method positions.str2pos(). This method is tested with script mu_insidetest.py which is part of the examples tar file. |
| Examples: | >>> fitsobj = maputils.FITSimage("m101.fits")
>>> fitsobj.set_limits((180,344), (100,200))
>>> annim = fitsobj.Annotatedimage()
>>> pos="{} 210.870170 {} 54.269001"
>>> print annim.inside(pos=pos)
>>> pos="ga 101.973853, ga 59.816461"
>>> print annim.inside(pos=pos)
>>> x = range(180,400,40)
>>> y = range(100,330,40)
>>> print annim.inside(x=x, y=y, mode='pixels')
>>> print annim.inside(x=crval1, y=crval2, mode='w')
|
Create a histogram equalized version of the data. The histogram equalized data is stored in attribute data_hist.
Blur the image by convolving with a gaussian kernel of typical size nx (pixels). The optional keyword argument ny allows for a different size in the y direction. nx, ny are the sigma’s for the gaussian kernel.
Allow this Annotatedimage object to interact with the user. It reacts to mouse movements. A message is prepared with position information in both pixel coordinates and world coordinates. The world coordinates are in the units given by the (FITS) header.
| Parameters: |
|
||
|---|---|---|---|
| Notes: | If a format is set to None, its corresponding number(s) will not appear in the informative message. If a message does not fit in the toolbar then only a part is displayed. We don’t have control over the maximum size of that message because it depends on the backend that is used (GTK, QT,...). If nothing appears, then a manual resize of the window will suffice. |
||
| Example: | Attach to an object from class Annotatedimage: >>> annim = f.Annotatedimage(frame)
>>> annim.interact_toolbarinfo()
or: >>> annim.interact_toolbarinfo(wcsfmt=None, zfmt="%g")
A more complete example:
|
Add mouse interaction (right mouse button) and keyboard interaction to change the colors in an image.
MOUSE
If you move the mouse in the image for which you did register this callback function and press the right mouse button at the same time, then the color limits for image and colorbar are set to a new value.
The new color setting is calculated as follows: first the position of the mouse (x, y) is transformed into normalized coordinates (i.e. between 0 and 1) called (xn, yn). These values are used to set the slope and offset for a function that sets an color for an image value according to the relations: slope = tan(89 * xn); offset = yn - 0.5. The minimum and maximum values of the image are set by parameters clipmin and clipmax. For a mouse position exactly in the center (xn,yn) = (0.5,0.5) the slope is 1.0 and the offset is 0.0 and the colors will be divided equally between clipmin and clipmax.
KEYBOARD
- page-down move forwards through a list with known color maps.
- page-up move backwards through a list with known color maps.
- 0 resets the colors to the original colormap and scaling. The default color map is ‘jet’.
- i (or ‘I’) toggles between inverse and normal scaling.
- 1 sets the colormap scaling to linear
- 2 sets the colormap scaling to logarithmic
- 3 sets the colormap scaling to exponential
- 4 sets the colormap scaling to square root
- 5 sets the colormap scaling to square
- b (or ‘B’) changes color of bad pixels.
- h (or ‘H’) replaces the current data by a histogram equalized version of this data. This key toggles between the original data and the equalized data.
- z (or ‘Z’) replaces the current data by a smoothed version of this data. This key is a toggle between the original data and the blurred version, smoothed with a value of sigma set by key ‘x’. Pressing ‘x’ repeatedly increases the smoothing factor. Note that Not a Number (NaN) values are smoothed to 0.0.
- x (or ‘X”) increases the smoothing factor. The number of steps is 10. Then is starts again with step 1.
- m (or ‘M’) saves current colormap look up data to a file. The default name of the file is the name of file from which the data was extracted or the name given in the constructor. The name is appended with ‘.lut’. This data is written in the right format so that it can be be (re)used as input colormap. This way you can fix a color setting and reproduce the same setting in another run of a program that allows one to enter a colormap from file.
If annim is an object from class Annotatedimage then activate color editing with:
1 2 3 4 5 6 7 | >>> fits = maputils.FITSimage("m101.fits")
>>> fig = plt.figure()
>>> frame = fig.add_subplot(1,1,1)
>>> annim = fits.Annotatedimage(frame)
>>> annim.Image()
>>> annim.interact_imagecolors()
>>> annim.plot()
|
Add mouse interaction (left mouse button) to write the position of the mouse to screen. The position is written both in pixel coordinates and world coordinates.
| Parameters: |
|
||
|---|---|---|---|
| Example: |
For a formatted output one could add parameters to interact_writepos(). The next line writes no pixel coordinates, writes spatial coordinates in degrees (not in HMS/DMS format) and adds a format for the world coordinates and the image value(s). >>> annim.interact_writepos(pixfmt=None, wcsfmt="%.12f", zfmt="%.3e",
hmsdms=False)
|
Read positions from a file with world coordinates and convert them to pixel coordinates. The interface is exactly the same as from method tabarray.readColumns()
It expects that the first column you specify contains the longitudes and the second column that is specified the latitudes.
| Parameters: |
|
|---|---|
| Examples: | >>> fn = 'smallworld.txt'
>>> xp, yp = annim.positionsfromfile(fn, 's', cols=[0,1])
>>> frame.plot(xp, yp, ',', color='#FFDAAA')
Or: your graticule is equatorial but the coordinates in the file are galactic: >>> xp, yp = annim.positionsfromfile(fn, 's', skyout='ga', cols=[0,1])
|
Prepare the FITS- or external image data to be plotted in Matplotlib. All parameters are set by method Annotatedimage.Image(). The keyword arguments are those for Matplotlib’s method imshow(). Two of them are useful in the context of this class. These parameters are visible, a boolean to set the visibility of the image to on or off, and alpha, a number between 0 and 1 which sets the transparency of the image.
See also: Annotatedimage.Image()
| Methods: |
|---|
Plot image object. Usually this is done by method Annotatedimage.plot() but it can also be used separately.
Objects from this class calculate and plot contour lines. Most of the parameters are set by method Annotatedimage.Contours(). The others are:
| Parameters: |
|
|---|---|
| Notes: | If the line widths of contours are given in the constructor (parameter linewidths) then these linewidths are copied to the line widths in the colorbar (if requested). |
| Methods: |
Plot contours object. Usually this is done by method Annotatedimage.plot() but it can also be used separately.
Set properties for contours either for all contours if levels is omitted or for specific levels if keyword levels is set to one or more levels.
| Parameters: | levels (None or one or a sequence of numbers) – None or one or more levels from the set of given contour levels |
|---|---|
| Examples: | >>> cont = annim.Contours(levels=range(10000,16000,1000))
>>> cont.setp_contour(linewidth=1)
>>> cont.setp_contour(levels=11000, color='g', linewidth=3)
|
Set properties for the labels along the contours. The properties are Matplotlib properties (fontsize, colors, inline, fmt).
| Parameters: |
|
|---|---|
| Examples: | >>> cont2 = annim.Contours(levels=(8000,9000,10000,11000))
>>> cont2.setp_label(11000, colors='b', fontsize=14, fmt="%.3f")
>>> cont2.setp_label(fontsize=10, fmt="%g \lambda")
|
Colorbar class. Usually the parameters will be provided by method Annotatedimage.Colorbar()
Useful keyword parameters:
| Parameters: | frame (Matplotlib Axes instance) – If a frame is given then this frame will be the colorbar frame. If None, the frame is calculated by taking space from its parent frame. |
|---|---|
| Methods: |
Plot image object. Usually this is done by method Annotatedimage.plot() but it can also be used separately.
Note: We changed the default formatter for the colorbar. This can be done with the ‘format’ parameter. We changed the formatter to a fixed format string. This prevents that MPL uses an offset and a scaling for the labels. If MPL does this, it adds an extra label, showing the offset and scale in scientific notation. We do not want this extra label because we don’t have enough control over it (e.g. it can appear outside your viewport or in a black background with a black font). With the new formatter we are sure to get the real value in our labels.
Set a text label along the long side of the color bar. It is a convenience routine for Matplotlib’s set_label() but this one needs a plotted colorbar while we postpone plotting.
Beam class. Usually the parameters will be provided by method Annotatedimage.Beam()
Objects from class Beam are graphical representations of the resolution of an instrument. The beam is centered at a position xc, yc. The major axis of the beam is the FWHM of longest distance between two opposite points on the ellipse. The angle between the major axis and the North is the position angle.
Note that it is not correct to calculate the ellipse that represents the beam by applying distance ‘r’ (see code) as a function of angle, to get the new world coordinates. The reason is that the fwhm’s are given as sizes on a sphere and therefore a correction for the declination is required. With method dispcoord() (see source code of class Beam) we sample the ellipse on a sphere with a correct position angle and with the correct sizes.
Note that a beam can also be specified in ‘pixel’/’grid’ units. Then the ellipse axes and center are in pixel coordinates, while the position angle is defined with respect to the positive X axis (i.e. not the astronomical position angle).
This class defines objects that can only be plotted onto spatial maps. Usually the parameters will be provided by method Annotatedimage.Skypolygon()
Marker class. Usually the parameters will be provided by method Annotatedimage.Marker()
Mark features in your map with a marker symbol. Properties of the marker are set with Matplotlib’s keyword arguments.
Draw positions in pixels along one or more plot axes. Nice numbers and step size are calculated by Matplotlib’s own plot methods.
| Parameters: |
|
||
|---|---|---|---|
| Returns: | An object from class Gridframe which is added to the plot container with Plotversion’s method Plotversion.add(). |
||
| Notes: | Graticules and Pixellabels are plotted in their own plot frame. If you want to be able to toggle grid lines in a frame labeled with pixel coordinates, then you have to make sure that the Pixellabels frame is plotted last. So always define Pixellabels objects before Graticule objects. |
||
| Examples: | Annotate the pixels in a plot along the right and top axis of a plot. Change the color of the labels to red:
|
||
| Methods: |
Set properties of the pixel label tick markers
| Parameters: | kwargs (Python keyword arguments.) – keyword arguments to change properties of the tick marks. A tick mark is a Matploltlib Line2D object with attributes like markeredgewidth etc. |
|---|
Set properties of the pixel label tick markers
| Parameters: | kwargs (Python keyword arguments.) – keyword arguments to change properties of (all) the tick labels. A tick mark is a Matploltlib Text object with attributes like fontsize, fontstyle etc. |
|---|
This class provides an object which stores the names of all available colormaps. The method add() adds external colormaps to this list. The class is used in the context of other classes but its attribute colormaps can be useful.
| Parameters: | - – No parameters |
|---|---|
| Attributes: |
|
This class defines objects which store WCS information from a FITS header. It includes axis number and alternate header information in a FITS keyword.
| Parameters: |
|
|---|---|
| Methods: |
Print formatted information for this axis.
| Examples: |
|---|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 | >>> from kapteyn import maputils
>>> fitsobject = maputils.FITSimage('rense.fits')
>>> fitsobject.hdr
<pyfits.NP_pyfits.Header instance at 0x1cae3170>
>>> ax1 = maputils.FITSaxis(1, fitsobject.hdr)
>>> ax1.printattr()
axisnr - Axis number: 1
axlen - Length of axis in pixels (NAXIS): 100
ctype - Type of axis (CTYPE): RA---NCP
axname - Short axis name: RA
cdelt - Pixel size: -0.007165998823
crpix - Reference pixel: 51.0
crval - World coordinate at reference pixel: -51.2820847959
cunit - Unit of world coordinate: DEGREE
wcstype - Axis type according to WCSLIB: None
wcsunits - Axis units according to WCSLIB: None
outsidepix - A position on an axis that does not belong to an image: None
|
If we set the image axes in fitsobject then the WCS attributes will get a value also. This object stores its FITSaxis objects in a list called axisinfo[]. The index is the required FITS axis number.
1 2 3 4 5 6 | >>> fitsobject.set_imageaxes(1, 2, 30)
>>> fitsobject.axisinfo[1].printattr()
..........
wcstype - Axis type according to WCSLIB: longitude
wcsunits - Axis units according to WCSLIB: deg
outsidepix - A position on an axis that does not belong to an image: None
|
Print formatted information for this axis.
| Examples: |
|
||
|---|---|---|---|
| Notes: | if attributes for a maputils.FITSimage object are changed then the relevant axis properties are updated. So this method can return different results depending on when it is used. |
This class creates an object with attributes that are needed to set a proper message with information about a position in a map and its corresponding image value. The world coordinates are calculated in the sky system of the image. This system could have been changed by the user.
The input parameters are usually set after initialization of an object from class Annotatedimage. For users/programmers the atributes are more important. With the attributes of objects of this class we can change the format of the numbers in the informative message.
Note that the methods of this class return separate strings for the pixel coordinates, the world coordinates and the image values. The final string is composed in the calling environment.
| Parameters: |
|
|---|---|
| Attributes: |
|
This class is a container for objects from class maputils.Annotatedimage. For this container there are methods to alter the visibility of the stored objects to get the effect of a movie loop. The objects are appended to a list with method maputils.MovieContainer.append(). With method MovieContainer.movie_events() the movie is started and keys ‘P’, ‘<’, ‘>’, ‘+’ and ‘-‘ are available to control the movie.
‘P’ : Pause/resume movie loop
‘<’ : Step 1 image back in the sequence of images. Key ‘,’ and arrow down has the same effect.
‘>’ : Step 1 image forward in the sequence of images. Key ‘.’ and arrow up has the same effect.
the hardware in use.
‘-‘ : Decrease the speed of the movie loop
One can also control the movie with method MovieContainer.controlpanel()
Usually one creates a movie container with class class:Cubes
| Parameters: |
|
||
|---|---|---|---|
| Attributes: |
|
||
| Examples: | Use of this class as a container for images in a movie loop:
Skip informative text on the display: >>> movieimages = maputils.MovieContainer(helptext=False, imageinfo=False)
|
||
| Methods: |
Append object from class Annotatedimage. First there is a check for the class of the incoming object. If it is the first object that is appended then from this object the Matplotlib figure instance is copied.
| Parameters: |
|
|---|---|
| Raises: | ‘Container object not of class maputils.Annotatedimage!’ An object was not recognized as a valid object to append. |
Connect keys for movie control and start the movie. Note that we defined the callbacks to trigger on the entire canvas. We don’t need to connect it to a frame (AxesCallback). We have to disconnect the callbacks explicitly.
Process the key and scroll events for the movie container. An external process (i.e. not the Matplotlib canvas) can access the control panel. It uses parameter ‘externalkey’. In the calling environment one needs to set ‘event’ to None.
Set a list with messages that will be used to display information when a frame is changed. This message is either printed on the image or returned, using a callback function, to the calling environment. List may be smaller than the number of images in the movie. A message should correspond to the movie index number.
Set a movie frame by index number. If the index is invalid, then do nothing.
| Parameters: | i – Index of movie frame which is any number between 0 and the number of loaded images. |
|---|
Helper method to get movie loop
| Parameters: | cb (Callback object based on matplotlib.backend_bases.MouseEvent instance) – Mouse event object with pixel position information. |
|---|
Toggle the visible state of images either by a timed callback function or by keys. This toggle works if one stacks multiple images in one frame with method MovieContainer.append(). Only one image gets status visible=True. The others are set to visible=False. This toggle changes this visibility for images and the effect, is a movie.
| Parameters: |
|
|---|
Class that sets cube attributes. The constructor is called by the maputils.Cubes.append() method of class maputils.Cubes and should not be used otherwise. We document the class because it has some useful attributes.
The attributes should only be used as a read-only attribute.
| Parameters: | xxx – All parameters are set by the maputils.Cubes class. |
|---|---|
| Attributes: |
|
A container with Cubeatts objects. With this class we build the movie container which can store images from different data cubes.
| Parameters: |
|
||
|---|---|---|---|
| Example: | Example 1 (Use callback functions in the constructor of a Cubes object)
|
||
| Example: | Use two different cube containers connected to two different figures
|
||
| Attributes: |
|
||
| Methods: |
Add a new cube to the container with cubes. Let’s introduce some definitions first:
- A data cube is a FITS file with more than two data axes. Examples are a sequence of channel maps in a radio cube where the spatial maps are a function of the observing freequency.
- Image axes are the two axes of a map we want to display Usually these maps have two spatial axes (e.g. R.A. and Dec.), but this class can also handle images which are slices with axes of mixed types (e.g. R.A. and velocity, a so called position velocity diagram).
- Repeat axes are all the axes that do not belong to the set of image axes. Along these repeat axes, one can define a sequence of images, for example a set of R.A., Dec. images as function of velocity or Stokes parameter.
| Parameters: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Examples: | 1) Simple example with a 3D data FITS file. The cube has axes RA, DEC and VELO. We show how to initialize a Cubes container and how to add objects of type maputils.FITSimage to this container:
2) List with callbacks. In this example we demonstrate how triggers for callbacks can be used. The use of Lambda expressions is demonstrated for functions that need extra parameters. If you run this example you will soon discover how these callbacks work:
|
Change aspect ratio of frame of ‘cube’.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Load a cube with images and set the pixel aspect ratio:
|
||
| Notes: | Note for programmers: We need a call to method maputils.Cubes.reposition() to adjust the graticule frames |
This method draws or removes a colorbar. Each cube is associated with a color bar.
A color bar will always be plotted to the left side of your plot and it have the same height as the canvas. The color bar will change its colours if you move the mouse while pressing the right mouse button. It will also change if you use color navigation with keyboard keys:
pgUp and pgDown: browse through colour maps
Right mouse button: Change slope and offset
h: Toggle histogram equalization & raw image
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Plot a color bar:
|
Set modes for the formatting of coordinate information. The output is generated by Annotatedimage.interact_writepos(). Coordinates are written to the terminal (by default) if the left mouse button is pressed simultaneously with the Shift button. The position corresponds to the position of the mouse cursor. For maps with one spatial axis, the coordinate of the matching spatial axis is also written.
There are several options to format the output. World coordinates follow the syntax of the system set by maputils.Cubes.set_skyout() and maputils.Cubes.set_spectrans().
Note that the settings in this method, only applies to the coordinates that are written to terminal (or log file). The labels on the graticule are not changed.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Modify the output of coordinates. We want a position written to a terminal in pixels, formatted and unformatted world coordinates and we also want the image value at the current cursor location:
|
This method prepares a crosshair cursor, which can be moved with the mouse while pressing the left mouse button.
Usually we want a crosshair cursor, to find the location of features. The crosshair cursor extends also to the side panels where the indicate at which slice we are looking.
Note that the situation with blitted images and multiple frames (Axes objects) prevents the use of class:matplolib.widgets.Cursor so an alternative is implemented.
| Parameters: | status (Boolean) – If set to True, a crosshair cursor will be plotted. If False, an existing crosshair will be removed. |
||
|---|---|---|---|
| Examples: | Draw a crosshair on the canvas which can be moved with the mouse while pressing the left mouse button. Extend the cursor in two side panels:
|
This method creates the WCS overlay and refreshes the current image or the relevant image if the current image does not belong to the cube for which a graticule is requested. The method can be used if the hasgraticule parameter in maputils.Cubes.append() has not been set.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Define a callback which is triggered after loading all the images in a data cube. The callback function extracts the first (and only) cube of a list of cubes and plots the graticules:
|
Set a list with movie frame index numbers to show up in one of the side panels. The panels appear when the list is not empty.
Imagine a 3D data structure. The axes of such a structure could be RA, DEC and FREQ. When we appended images, we specified the axis numbers in axnums. Assume these numbers are (1,2) which corresponds to images with axes RA and DEC. We can show a movie loop of the images as function of positions in axis FREQ. A side panel is a new image that shares either the RA or the DEC axis with the cube, but the other axis is a FREQ axis. This way we extracted a slice from the cube. panel='X' gives a horizontal RA-FREQ slice and panel='Y' gives a vertical DEC-FREQ slice and the data is extracted at the position of the cursor in the current image and when you move the mouse while pressing the left mouse button, you will see that the side panels are updated with new data extracted from the new mouse cursor position. It is also possible use the mouse in the side panels. If you move the mouse in the FREQ direction, you will see that the current image is replaced by the one that matches the new slice position in FREQ. If you move in a side panel in the RA or DEC direction, then you will see that the opposite side panel is updated because the position where the slice was extracted (DEC or RA) has been changed. This functionality has been used often for the inspection of radio data because it allows you to discover features in the data in a simple and straightforward way.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Set both side panels to inspect orthogonal slices:
|
This method changes the celestial system. It redraws the graticule so that its labels represent the new celestial system.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Change the sky system from the native system to Galactic and draw a graticule showing the world coordinate system:
|
This method changes the spectral system. If a valid system is defined, you will notice that spectral coordinates are printed in the new system. If a graticule has a spectral axis, you will see that the labels will change so that they represent the new spectral system.
| Parameters: |
|
||
|---|---|---|---|
| Examples: | Change the sky system from the native spectral system to wavelengths (in m) and draw a graticule showing the world coordinate system. Note that we loaded images with one spatial and one spectral axis (the cube has a RA, DEC and VELO axis) with axnums=(1,3). To get a list with valid spectral translations, we printed fitsobject.allowedtrans:
|
Set image (by its number) which is used to split the current screen.
| Parameters: | nr (Integer) – Number of the movie frame that corresponds to the image you want to use together with the current image to split and compare. Note that these movieframe numbers start with 0 and run to the number of loaded frames. The numbering continues after loading more than 1 data cube. |
|---|
This method plots (or removes) a so called z-profile as a (non-filled) histogram as function of cursor position in an image.
The data is extracted from a data set, with at least three axes, at the cursor position in the current image and this is repeated for all the images in all the cubes that were entered. This is also repeated for all the repeat axex that you specified for a given cube. So the X axis of the z-profile is the movie frame number and not a slice number. The z-profile is shown per cube. If you change to an image that belongs to another cube, the z-profile will change accordingly, but the X axis still represents the movie frame number.
You can update the z-profile by moving the mouse while pressing the left mouse button. Change the limits in the Y direction (image values) with method maputils.Cubes.zprofileLimits().
| Parameters: | status (Boolean) – Plot z-profile if status=True. If there is already a z-profile plotted, then remove it with status=False |
||
|---|---|---|---|
| Examples: | Load a cube and plot its z-profile. Set the range of image values for the z-profile:
|
Set limits of plot with Z_profile for the current cube.
| Parameters: |
|
|---|---|
| Notes: | See example at maputils.Cubes.set_zprofile() |